Cindy's DeCAF paper on detecting cell-type specific QTLs with allele-specific expression is out (Kalita et al. Genom Biol)! 🎉
Work led by Cynthia Kalita, developing a new method to identify cell-type specific QTLs in bulk RNA-seq data by leveraging allele-specificity is now out in Genome Biology:
DeCAF: A novel method to identify cell-type specific regulatory variants and their role in cancer risk.
Kalita C, Gusev A. Genome Biology. 2022
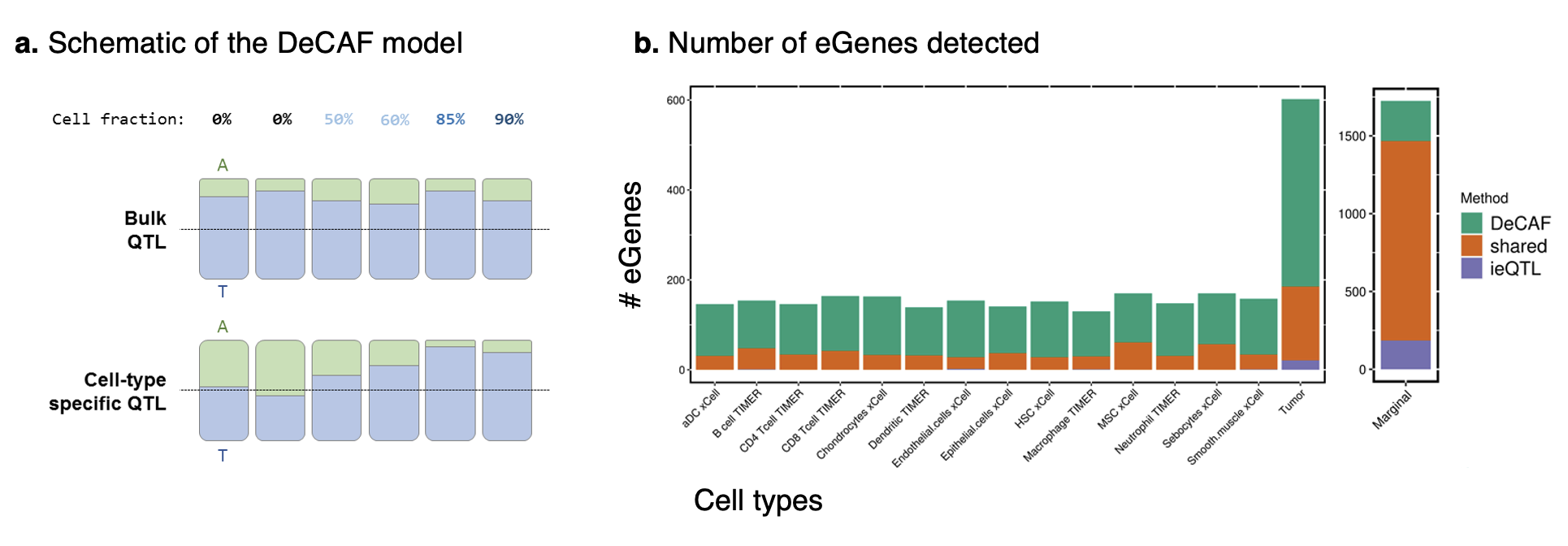
Several recent studies have shown that variants influencing gene expression in a cell-type specific manner can be detected from bulk (i.e. non-specific) RNA-seq data by leveraging differences in cell type proportions across individuals. Such methods typically work by modeling the interaction between the genotypic effect on expression and individual cell type proportions: higher effects in individuals with more of a given cell type provide evidence of specificity. Here, we propose the method DeCAF to additionally incorporate allele-specific expression: the difference in expression between the two haplotypes of an individual. Individuals with more of a given cell type are expected to have more allele-specificity in their expression, which is a source of signal that is independent and complementary to the conventional eQTL interaction.
In simulation and in real data, DeCAF greatly boosts power to detect cell-type specific effects at low sample sizes. Surprisingly, we find the largest number of such effects coming from the tumor (where tumor purity is treated as “cell type” proportion). These tumor-specific effects replicate strongly in immune cell types, suggesting they may be capturing genetic variants involved in tumor-immune interactions.
DeCAF can be broadly applied to cancer and non-cancer studies to identify additional interesting QTLs, and is open source.
Figure: Summary of the DeCAF model and results