Doug's paper on efficient recovery of effects from perturbational screens (Yao et al. Nature Biotech) 🎉
Work led by Doug Yao on using compressed sensing to efficiently identify effects in composite perturbational screens and link these effects to disease heritability is now out in Nature Biotechnology. This was a collaboration with the Regev and Cleary labs:
Scalable genetic screening for regulatory circuits using compressed Perturb-seq
D Yao, L Binan, J Bezney, B Simonton, J Freedman, CJ Frangieh, K Dey, K Geiger-Schuller, B Eraslan, A Gusev[+], A Regev[+], Brian Cleary[+]
Nature Biotechnology. 2023
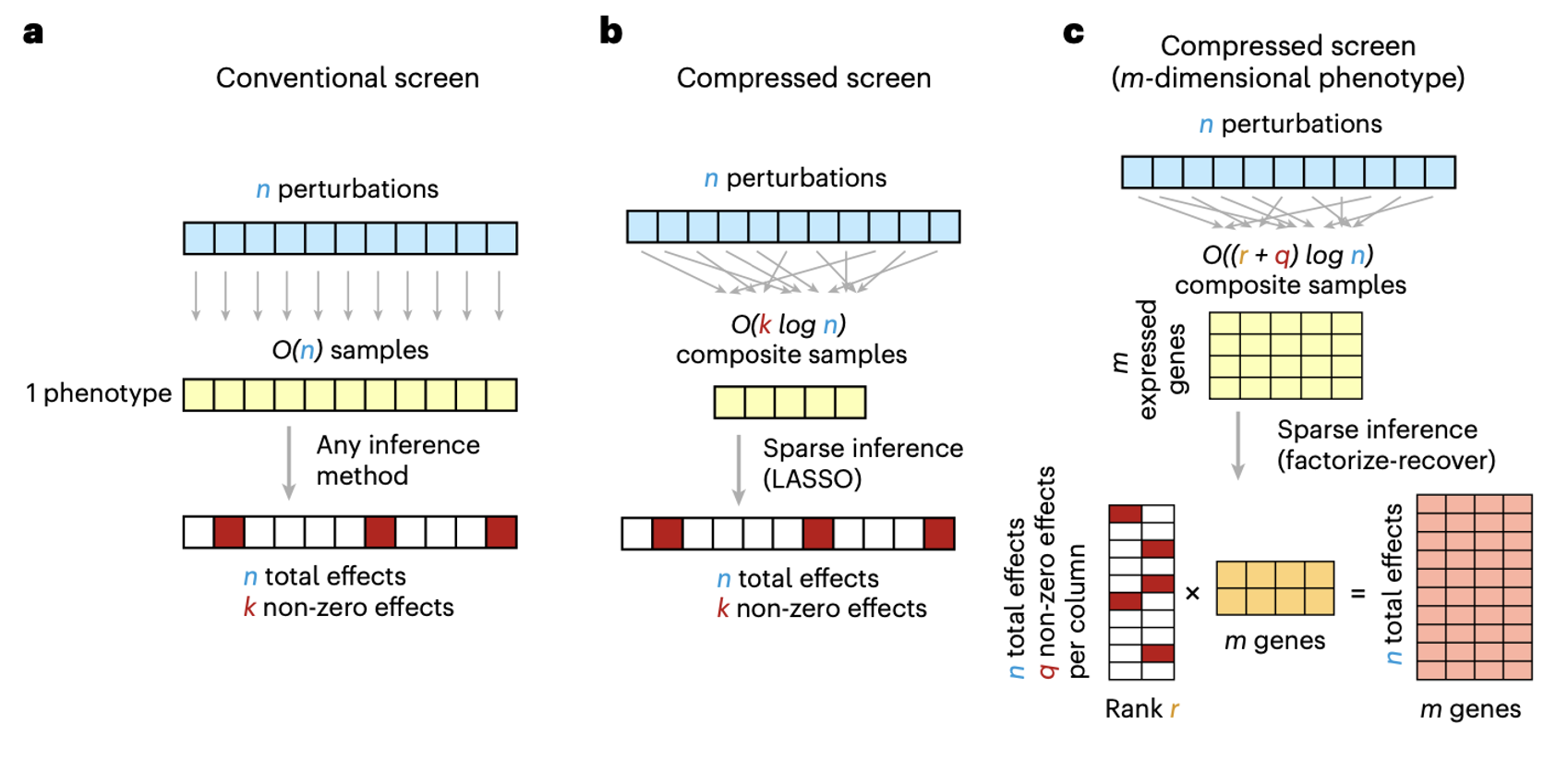
Perturb-seq is a technology for evaluating the effect of many perturbations (gene knock-down/knock-out) on the expression of all other genes in a single-cell assay, however it typically requires a very large number of cells. In this work, a “compressed” approach was proposed where multiple perturbations are induced into each cell (or cell droplet) simultaneously and then their individual effects are “recovered” using an algorithm based on compressed sensing. The paper showed that this approach can increase the efficiency by 4-20x relative to a conventional screen while retaining the same accuracy. The approach also potentially enables the recover of genetic interactions. Finally, the relationship between perturbational effects, GWAS disease heritability, and population-scale QTL effects were investigated with intriguing findings.
For more, see the Research Briefing and thread from Doug. The software is open source and on github.
Schematic of the (a) conventional screen, (b) novel compressed screen, and (c) novel algorithmic recovery.